Step-1: A first spatial problem¶
In this step, we create a spatial problem and play with it a little bit. No SDC, no multi-level here so far.
Part A: Spatial problem setup¶
We start as simple as possible by creating a first, very simple problem
using one of the problem_classes, namely HeatEquation_1D_FD.
This basically consists only of the matrix A, which represents a
finite difference discretization of the 1D Laplacian. This is tested for
one particular example.
Important things to note:
Many (most) parameters for pySDC are passed using dictionaries
Data types are encapsulated: the real values are stored in
values, meta-information can be stored separately in the data structureA quick peak into
HeatEquation_1D_FDreveals that theinitand theparams.nvarsattribute contain the same values (namelynvars). Yet, sometimes one or the other is used here (and throughout the code). The reason is this: the data structure (meshin this case) needs some form of standard initialization. For this, pySDC uses theinitattribute each problem class has. In our simple example, this is he same asnvars, but e.g. for Finite Elements, the function space is stored in theinitattribute. Thus, whenever a data type is created, useinit, otherwise do whatever looks nice.We happily pass classes around so that they can be used to instantiate things within subroutines
Full code: pySDC/tutorial/step_1/A_spatial_problem_setup.py
import numpy as np
from pathlib import Path
from pySDC.implementations.problem_classes.HeatEquation_ND_FD import heatNd_unforced
def main():
"""
A simple test program to set up a spatial problem and play with it
"""
# instantiate problem
prob = heatNd_unforced(
nvars=1023, # number of degrees of freedom
nu=0.1, # diffusion coefficient
freq=4, # frequency for the test value
bc='dirichlet-zero', # boundary conditions
)
# run accuracy test, get error back
err = run_accuracy_check(prob)
Path("data").mkdir(parents=True, exist_ok=True)
f = open('data/step_1_A_out.txt', 'w')
out = 'Error of the spatial accuracy test: %8.6e' % err
f.write(out)
print(out)
f.close()
assert err <= 2e-04, "ERROR: the spatial accuracy is higher than expected, got %s" % err
def run_accuracy_check(prob):
"""
Routine to check the error of the Laplacian vs. its FD discretization
Args:
prob: a problem instance
Returns:
the error between the analytic Laplacian and the computed one of a given function
"""
# create x values, use only inner points
xvalues = np.array([(i + 1) * prob.dx for i in range(prob.nvars[0])])
# create a mesh instance and fill it with a sine wave
u = prob.dtype_u(init=prob.init)
u[:] = np.sin(np.pi * prob.freq[0] * xvalues)
# create a mesh instance and fill it with the Laplacian of the sine wave
u_lap = prob.dtype_u(init=prob.init)
u_lap[:] = -((np.pi * prob.freq[0]) ** 2) * prob.nu * np.sin(np.pi * prob.freq[0] * xvalues)
# compare analytic and computed solution using the eval_f routine of the problem class
err = abs(prob.eval_f(u, 0) - u_lap)
return err
if __name__ == "__main__":
main()
Results:
Error of the spatial accuracy test: 1.981783e-04
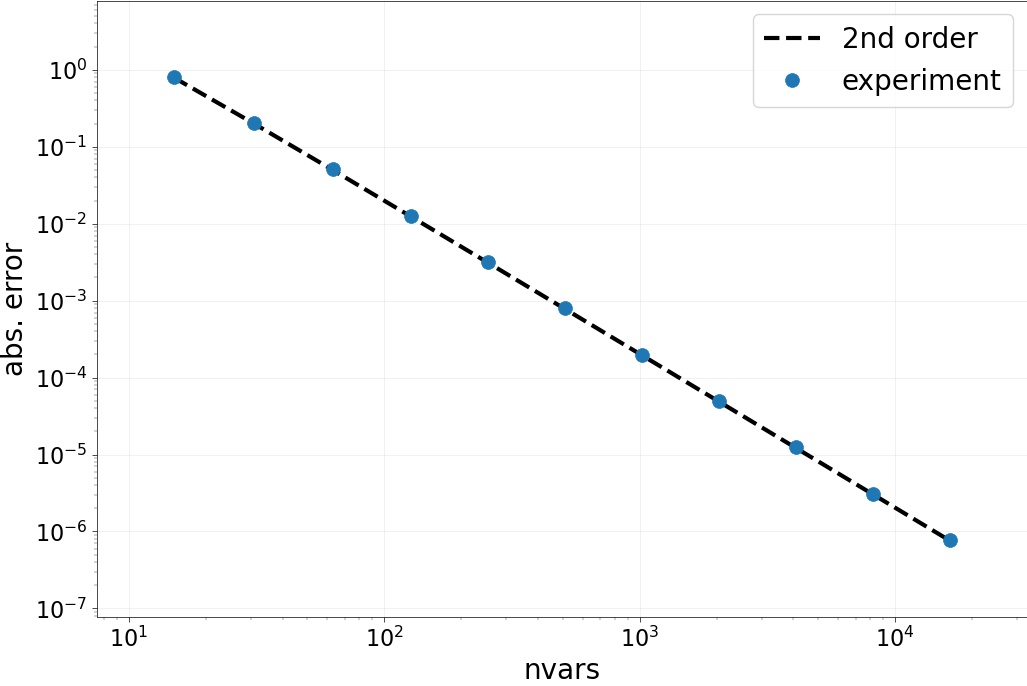
Part B: Spatial accuracy check¶
We now do a more thorough test of the accuracy of the spatial
discretization. We loop over a set of nvars, compute the Laplacian
of our test vector and look at the error. Then, the order of accuracy in
space is checked by looking at the numbers and by looking at “points on a line”.
Important things to note:
You better test your operators.. use nvars > 2**16 and things will break!
Add your results into a dictionary for later usage. Use IDs to find the data! Also, use headers to store meta-information.
Full code: pySDC/tutorial/step_1/B_spatial_accuracy_check.py
from pathlib import Path
import matplotlib
matplotlib.use('Agg')
from collections import namedtuple
import matplotlib.pylab as plt
import numpy as np
import os.path
from pySDC.implementations.datatype_classes.mesh import mesh
from pySDC.implementations.problem_classes.HeatEquation_ND_FD import heatNd_unforced
# setup id for gathering the results (will sort by nvars)
ID = namedtuple('ID', 'nvars')
def main():
"""
A simple test program to check order of accuracy in space for a simple test problem
"""
# initialize problem parameters
problem_params = dict()
problem_params['nu'] = 0.1 # diffusion coefficient
problem_params['freq'] = 4 # frequency for the test value
problem_params['bc'] = 'dirichlet-zero' # boundary conditions
# create list of nvars to do the accuracy test with
nvars_list = [2**p - 1 for p in range(4, 15)]
# run accuracy test for all nvars
results = run_accuracy_check(nvars_list=nvars_list, problem_params=problem_params)
# compute order of accuracy
order = get_accuracy_order(results)
Path("data").mkdir(parents=True, exist_ok=True)
f = open('data/step_1_B_out.txt', 'w')
for l in range(len(order)):
out = 'Expected order: %2i -- Computed order %4.3f' % (2, order[l])
f.write(out + '\n')
print(out)
f.close()
# visualize results
plot_accuracy(results)
assert os.path.isfile('data/step_1_accuracy_test_space.png'), 'ERROR: plotting did not create file'
assert all(np.isclose(order, 2, rtol=0.06)), "ERROR: spatial order of accuracy is not as expected, got %s" % order
def run_accuracy_check(nvars_list, problem_params):
"""
Routine to check the error of the Laplacian vs. its FD discretization
Args:
nvars_list: list of nvars to do the testing with
problem_params: dictionary containing the problem-dependent parameters
Returns:
a dictionary containing the errors and a header (with nvars_list)
"""
results = {}
# loop over all nvars
for nvars in nvars_list:
# setup problem
problem_params['nvars'] = nvars
prob = heatNd_unforced(**problem_params)
# create x values, use only inner points
xvalues = np.array([(i + 1) * prob.dx for i in range(prob.nvars[0])])
# create a mesh instance and fill it with a sine wave
u = prob.u_exact(t=0)
# create a mesh instance and fill it with the Laplacian of the sine wave
u_lap = prob.dtype_u(init=prob.init)
u_lap[:] = -((np.pi * prob.freq[0]) ** 2) * prob.nu * np.sin(np.pi * prob.freq[0] * xvalues)
# compare analytic and computed solution using the eval_f routine of the problem class
err = abs(prob.eval_f(u, 0) - u_lap)
# get id for this nvars and put error into dictionary
id = ID(nvars=nvars)
results[id] = err
# add nvars_list to dictionary for easier access later on
results['nvars_list'] = nvars_list
return results
def get_accuracy_order(results):
"""
Routine to compute the order of accuracy in space
Args:
results: the dictionary containing the errors
Returns:
the list of orders
"""
# retrieve the list of nvars from results
assert 'nvars_list' in results, 'ERROR: expecting the list of nvars in the results dictionary'
nvars_list = sorted(results['nvars_list'])
order = []
# loop over two consecutive errors/nvars pairs
for i in range(1, len(nvars_list)):
# get ids
id = ID(nvars=nvars_list[i])
id_prev = ID(nvars=nvars_list[i - 1])
# compute order as log(prev_error/this_error)/log(this_nvars/old_nvars) <-- depends on the sorting of the list!
tmp = np.log(results[id_prev] / results[id]) / np.log(nvars_list[i] / nvars_list[i - 1])
order.append(tmp)
return order
def plot_accuracy(results):
"""
Routine to visualize the errors as well as the expected errors
Args:
results: the dictionary containing the errors
"""
# retrieve the list of nvars from results
assert 'nvars_list' in results, 'ERROR: expecting the list of nvars in the results dictionary'
nvars_list = sorted(results['nvars_list'])
# Set up plotting parameters
params = {
'legend.fontsize': 20,
'figure.figsize': (12, 8),
'axes.labelsize': 20,
'axes.titlesize': 20,
'xtick.labelsize': 16,
'ytick.labelsize': 16,
'lines.linewidth': 3,
}
plt.rcParams.update(params)
# create new figure
plt.figure()
# take x-axis limits from nvars_list + some spacing left and right
plt.xlim([min(nvars_list) / 2, max(nvars_list) * 2])
plt.xlabel('nvars')
plt.ylabel('abs. error')
plt.grid()
# get guide for the order of accuracy, i.e. the errors to expect
# get error for first entry in nvars_list
id = ID(nvars=nvars_list[0])
base_error = results[id]
# assemble optimal errors for 2nd order method and plot
order_guide_space = [base_error / (2 ** (2 * i)) for i in range(0, len(nvars_list))]
plt.loglog(nvars_list, order_guide_space, color='k', ls='--', label='2nd order')
min_err = 1e99
max_err = 0e00
err_list = []
# loop over nvars, get errors and find min/max error for y-axis limits
for nvars in nvars_list:
id = ID(nvars=nvars)
err = results[id]
min_err = min(err, min_err)
max_err = max(err, max_err)
err_list.append(err)
plt.loglog(nvars_list, err_list, ls=' ', marker='o', markersize=10, label='experiment')
# adjust y-axis limits, add legend
plt.ylim([min_err / 10, max_err * 10])
plt.legend(loc=1, ncol=1, numpoints=1)
# save plot as PDF, beautify
fname = 'data/step_1_accuracy_test_space.png'
plt.savefig(fname, bbox_inches='tight')
return None
if __name__ == "__main__":
main()
Results:
Expected order: 2 -- Computed order 1.888
Expected order: 2 -- Computed order 1.949
Expected order: 2 -- Computed order 1.976
Expected order: 2 -- Computed order 1.988
Expected order: 2 -- Computed order 1.994
Expected order: 2 -- Computed order 1.997
Expected order: 2 -- Computed order 1.999
Expected order: 2 -- Computed order 1.999
Expected order: 2 -- Computed order 1.999
Expected order: 2 -- Computed order 1.982
Part C: Collocation problem setup¶
Here, we set up our first collocation problem using one of the
collocation_classes, namely GaussRadau_Right. Using the spatial
operator, we can build the collocation problem which in this case is
linear. This fully coupled system is then solved directly and the error
is compared to the exact solution.
Important things to note:
The collocation matrix
Qis and will be always relative to the temporal domain [0,1]. Usedtto scale it appropriately.Although convenient to analyze, the matrix formulation is not suited for larger (in space or time) computations. All remaining computations in pySDC are thus based on decoupling space and time operators (i.e. no Kronecker product).
We can use the
u_exactroutine here to return the values at any given point in time. It is recommended to include either an initialization routine or the exact solution (if applicable) into the problem class.This is where the fun with parameters comes in: How many DOFs do we need in space? How large/small does dt have to be? What frequency in space is fine? …
Full code: pySDC/tutorial/step_1/C_collocation_problem_setup.py
import numpy as np
import scipy.sparse as sp
from pathlib import Path
from pySDC.core.collocation import CollBase
from pySDC.implementations.problem_classes.HeatEquation_ND_FD import heatNd_unforced
def main():
"""
A simple test program to create and solve a collocation problem directly
"""
# instantiate problem
prob = heatNd_unforced(
nvars=1023, # number of degrees of freedom
nu=0.1, # diffusion coefficient
freq=4, # frequency for the test value
bc='dirichlet-zero', # boundary conditions
)
# instantiate collocation class, relative to the time interval [0,1]
coll = CollBase(num_nodes=3, tleft=0, tright=1, node_type='LEGENDRE', quad_type='RADAU-RIGHT')
# set time-step size (warning: the collocation matrices are relative to [0,1], see above)
dt = 0.1
# solve collocation problem
err = solve_collocation_problem(prob=prob, coll=coll, dt=dt)
Path("data").mkdir(parents=True, exist_ok=True)
f = open('data/step_1_C_out.txt', 'w')
out = 'Error of the collocation problem: %8.6e' % err
f.write(out + '\n')
print(out)
f.close()
assert err <= 4e-04, "ERROR: did not get collocation error as expected, got %s" % err
def solve_collocation_problem(prob, coll, dt):
"""
Routine to build and solve the linear collocation problem
Args:
prob: a problem instance
coll: a collocation instance
dt: time-step size
Return:
the analytic error of the solved collocation problem
"""
# shrink collocation matrix: first line and column deals with initial value, not needed here
Q = coll.Qmat[1:, 1:]
# build system matrix M of collocation problem
M = sp.eye(prob.nvars[0] * coll.num_nodes) - dt * sp.kron(Q, prob.A)
# get initial value at t0 = 0
u0 = prob.u_exact(t=0)
# fill in u0-vector as right-hand side for the collocation problem
u0_coll = np.kron(np.ones(coll.num_nodes), u0)
# get exact solution at Tend = dt
uend = prob.u_exact(t=dt)
# solve collocation problem directly
u_coll = sp.linalg.spsolve(M, u0_coll)
# compute error
err = np.linalg.norm(u_coll[-prob.nvars[0] :] - uend, np.inf)
return err
if __name__ == "__main__":
main()
Results:
Error of the collocation problem: 3.803471e-04
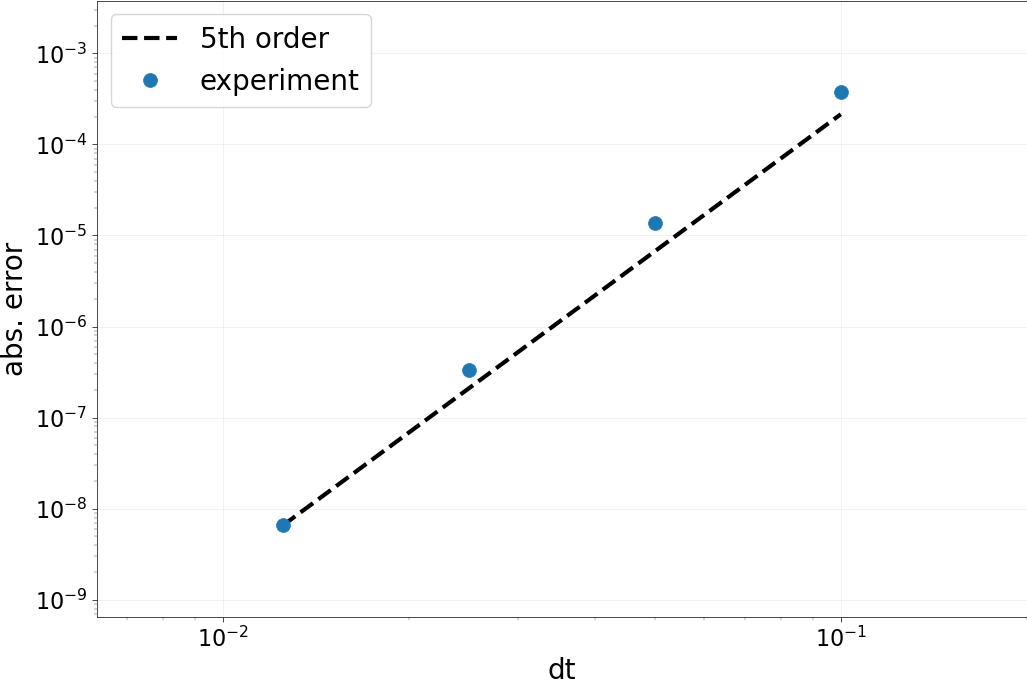
Part D: Collocation accuracy test¶
As for the spatial operator, we now test the accuracy in time. This
time, we loop over a set of dt, compute the solution to the
collocation problem and look at the error.
Important things to note:
We take a large number of DOFs in space, since we need to beat 5th order in time with a 2nd order stencil in space.
Orders of convergence are not as stable as for the space-only test. One of the problems of this example is that we are actually trying to compute 0 very, very thoroughly…
Full code: pySDC/tutorial/step_1/D_collocation_accuracy_check.py
from pathlib import Path
import matplotlib
matplotlib.use('Agg')
from collections import namedtuple
import matplotlib.pylab as plt
import numpy as np
import os.path
import scipy.sparse as sp
from pySDC.core.collocation import CollBase
from pySDC.implementations.problem_classes.HeatEquation_ND_FD import heatNd_unforced
# setup id for gathering the results (will sort by dt)
ID = namedtuple('ID', 'dt')
def main():
"""
A simple test program to compute the order of accuracy in time
"""
# initialize problem parameters
problem_params = {}
problem_params['nu'] = 0.1 # diffusion coefficient
problem_params['freq'] = 4 # frequency for the test value
problem_params['nvars'] = 16383 # number of DOFs in space
problem_params['bc'] = 'dirichlet-zero' # boundary conditions
# instantiate problem
prob = heatNd_unforced(**problem_params)
# instantiate collocation class, relative to the time interval [0,1]
coll = CollBase(num_nodes=3, tleft=0, tright=1, node_type='LEGENDRE', quad_type='RADAU-RIGHT')
# assemble list of dt
dt_list = [0.1 / 2**p for p in range(0, 4)]
# run accuracy test for all dt
results = run_accuracy_check(prob=prob, coll=coll, dt_list=dt_list)
# get order of accuracy
order = get_accuracy_order(results)
Path("data").mkdir(parents=True, exist_ok=True)
f = open('data/step_1_D_out.txt', 'w')
for l in range(len(order)):
out = 'Expected order: %2i -- Computed order %4.3f' % (5, order[l])
f.write(out + '\n')
print(out)
f.close()
# visualize results
plot_accuracy(results)
assert os.path.isfile('data/step_1_accuracy_test_coll.png')
assert all(np.isclose(order, 2 * coll.num_nodes - 1, rtol=0.4)), (
"ERROR: did not get order of accuracy as expected, got %s" % order
)
def run_accuracy_check(prob, coll, dt_list):
"""
Routine to build and solve the linear collocation problem
Args:
prob: a problem instance
coll: a collocation instance
dt_list: list of time-step sizes
Return:
the analytic error of the solved collocation problem
"""
results = {}
# loop over all nvars
for dt in dt_list:
# shrink collocation matrix: first line and column deals with initial value, not needed here
Q = coll.Qmat[1:, 1:]
# build system matrix M of collocation problem
M = sp.eye(prob.nvars[0] * coll.num_nodes) - dt * sp.kron(Q, prob.A)
# get initial value at t0 = 0
u0 = prob.u_exact(t=0)
# fill in u0-vector as right-hand side for the collocation problem
u0_coll = np.kron(np.ones(coll.num_nodes), u0)
# get exact solution at Tend = dt
uend = prob.u_exact(t=dt)
# solve collocation problem directly
u_coll = sp.linalg.spsolve(M, u0_coll)
# compute error
err = np.linalg.norm(u_coll[-prob.nvars[0] :] - uend, np.inf)
# get id for this dt and store error in results
id = ID(dt=dt)
results[id] = err
# add list of dt to results for easier access
results['dt_list'] = dt_list
return results
def get_accuracy_order(results):
"""
Routine to compute the order of accuracy in time
Args:
results: the dictionary containing the errors
Returns:
the list of orders
"""
# retrieve the list of dt from results
assert 'dt_list' in results, 'ERROR: expecting the list of dt in the results dictionary'
dt_list = sorted(results['dt_list'], reverse=True)
order = []
# loop over two consecutive errors/dt pairs
for i in range(1, len(dt_list)):
# get ids
id = ID(dt=dt_list[i])
id_prev = ID(dt=dt_list[i - 1])
# compute order as log(prev_error/this_error)/log(this_dt/old_dt) <-- depends on the sorting of the list!
tmp = np.log(results[id] / results[id_prev]) / np.log(dt_list[i] / dt_list[i - 1])
order.append(tmp)
return order
def plot_accuracy(results):
"""
Routine to visualize the errors as well as the expected errors
Args:
results: the dictionary containing the errors
"""
# retrieve the list of nvars from results
assert 'dt_list' in results, 'ERROR: expecting the list of dts in the results dictionary'
dt_list = sorted(results['dt_list'])
# Set up plotting parameters
params = {
'legend.fontsize': 20,
'figure.figsize': (12, 8),
'axes.labelsize': 20,
'axes.titlesize': 20,
'xtick.labelsize': 16,
'ytick.labelsize': 16,
'lines.linewidth': 3,
}
plt.rcParams.update(params)
# create new figure
plt.figure()
# take x-axis limits from nvars_list + some spacning left and right
plt.xlim([min(dt_list) / 2, max(dt_list) * 2])
plt.xlabel('dt')
plt.ylabel('abs. error')
plt.grid()
# get guide for the order of accuracy, i.e. the errors to expect
# get error for first entry in nvars_list
id = ID(dt=dt_list[0])
base_error = results[id]
# assemble optimal errors for 5th order method and plot
order_guide_space = [base_error * (2 ** (5 * i)) for i in range(0, len(dt_list))]
plt.loglog(dt_list, order_guide_space, color='k', ls='--', label='5th order')
min_err = 1e99
max_err = 0e00
err_list = []
# loop over nvars, get errors and find min/max error for y-axis limits
for dt in dt_list:
id = ID(dt=dt)
err = results[id]
min_err = min(err, min_err)
max_err = max(err, max_err)
err_list.append(err)
plt.loglog(dt_list, err_list, ls=' ', marker='o', markersize=10, label='experiment')
# adjust y-axis limits, add legend
plt.ylim([min_err / 10, max_err * 10])
plt.legend(loc=2, ncol=1, numpoints=1)
# save plot as PDF, beautify
fname = 'data/step_1_accuracy_test_coll.png'
plt.savefig(fname, bbox_inches='tight')
return None
if __name__ == "__main__":
main()
Results:
Expected order: 5 -- Computed order 4.791
Expected order: 5 -- Computed order 5.364
Expected order: 5 -- Computed order 5.675